본문
Modulation of mRNA and lncRNA expression dynamics by the Set2–Rpd3S pathway
by Prof. TaeSoo Kim (tskim@ewha.ac.kr)
Eukaryotic transcription is regulated by post-translational modifications of histones, including acetylation, methylation, phosphorylation and ubiquitination. Histone acetylation can promote RNA polymerase II (RNApII) transcription by disrupting the interaction between histone proteins and DNA or by recruiting factors that regulate chromatin structure and transcription. Histone acetylation is a highly dynamic modification regulated by both histone acetyltransferases and histone deacetylases (HDACs). Site-specific histone methylations also play important roles in regulating histone acetylation levels by targeting histone acetyltransferases or HDACs to specific gene locations. For example, H3K36 methylation by Set2 targets Rpd3S histone deacetylase to transcribed regions of mRNA genes, repressing internal cryptic promoters and slowing elongation.
The Set2 methyltransferase directly binds elongating RNApII and co-transcriptionally methylates H3K36. In early stages of transcription, the basal transcription factor TFIIH phosphorylates serine 5 of the C-terminal domain (CTD) of Rpb1, the largest subunit of RNApII, creating binding sites for factors involved in transcription initiation, histone modification and early termination. During transcription elongation, Ctk1 kinase phosphorylates serine 2 of CTD to create a binding site for the Set2 methyltransferase. In general, H3K36me2 and H3K36me3 are enriched in transcribed regions with increased levels towards 3’ ends, and this pattern is highly conserved in all eukaryotes. Methylated histone tails primarily function as binding sites for downstream effector proteins that have chromodomains, tudor domains or Plant Homeodomain (PHD) fingers. Nucleosomes methylated on H3K36 are deacetylated by the Rpd3 small complex, Rpd3S. The Eaf3 chromodomain and the Rco1 PHD finger mediate binding of Rpd3S to histones and this binding is stimulated by H3K36 methylation. Although Rpd3S may be in part recruited to transcribed regions via interaction with the phosphorylated CTD of RNApII, histone deacetylation by this complex requires H3K36 methylation. Deacetylation by the Set2–Rpd3S pathway represses transcription from cryptic internal promoters and slows transcription elongation.
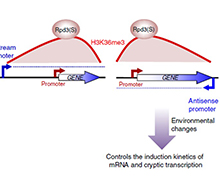
However, a recent study showed that the Set2–Rpd3S pathway had little effect on mRNA levels under steady-state laboratory growth conditions. We explore the function of this pathway by analysing transcription in yeast undergoing a series of carbon source shifts. Approximately 80 mRNA genes show increased induction upon SET2 deletion. A majority of these promoters have overlapping long non-coding RNA (lncRNA) transcription that targets H3K36me3 and deacetylation by Rpd3S to the mRNA promoter (Figure 1). We previously reported a similar mechanism for H3K4me2-mediated repression via recruitment of the Set3C HDAC. We show that the distance between an mRNA and overlapping lncRNA promoter determines whether Set2–Rpd3S or Set3C represses. This analysis also reveals many previously unreported cryptic ncRNAs induced by specific carbon sources, showing that cryptic promoters can be environmentally regulated (Figure 1). Therefore, in addition to repression of cryptic transcription and modulation of elongation, H3K36 methylation maintains optimal expression dynamics of many mRNAs and ncRNAs. This type of kinetic regulation is likely to be critically important for cells growing in the wild, where they must continuously alter gene expression patterns to adapt to changing environmental conditions.
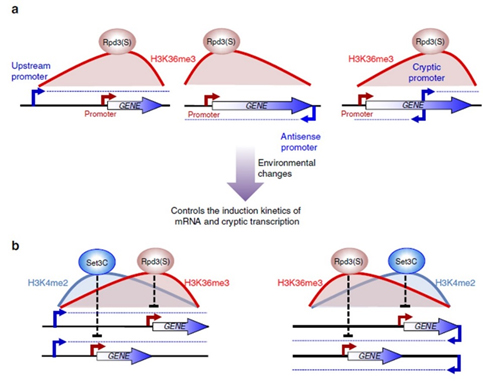
Figure 1. Models for regulation of gene expression by Set2–Rpd3S pathway and overlapping lncRNA transcription.
* Related Article
Modulation of mRNA and lncRNA expression dynamics by the Set2–Rpd3S pathway, 20161128, NATURE COMMUNICATIONS 7, 13534