본문
Drug Synergy Prediction Using Graph-Based Deep Learning

by Prof. Sang-Hyuk Lee
Department of Life Sciences
PURE Research Profile
sanghyuk@ewha.ac.kr
Objective
Drug combination therapy is a promising approach for developing new treatments, and various models have been created to suggest potential drug pairs. While the DrugComb database provides experimental results from high-throughput technologies, testing all possible drug combinations is impractical due to the large number of combinations. As a result, computational methods, including machine learning and graph convolutional networks (GCNs), have emerged as cost-effective tools for predicting drug synergy. These methods utilize molecular interaction data, such as drug-target interactions, protein-protein interactions, and gene expression profiles from resources like CCLE and LINCS to model the effects of drug combinations.
Overview
Drug combination therapy is a promising approach for developing new treatments, and various models have been created to suggest potential drug pairs. While the DrugComb database provides experimental results from high-throughput technologies, testing all possible drug combinations is impractical due to the large number of combinations. As a result, computational methods, including machine learning and graph convolutional networks (GCNs), have emerged as cost-effective tools for predicting drug synergy. These methods utilize molecular interaction data, such as drug-target interactions, protein-protein interactions, and gene expression profiles from resources like CCLE and LINCS to model the effects of drug combinations.
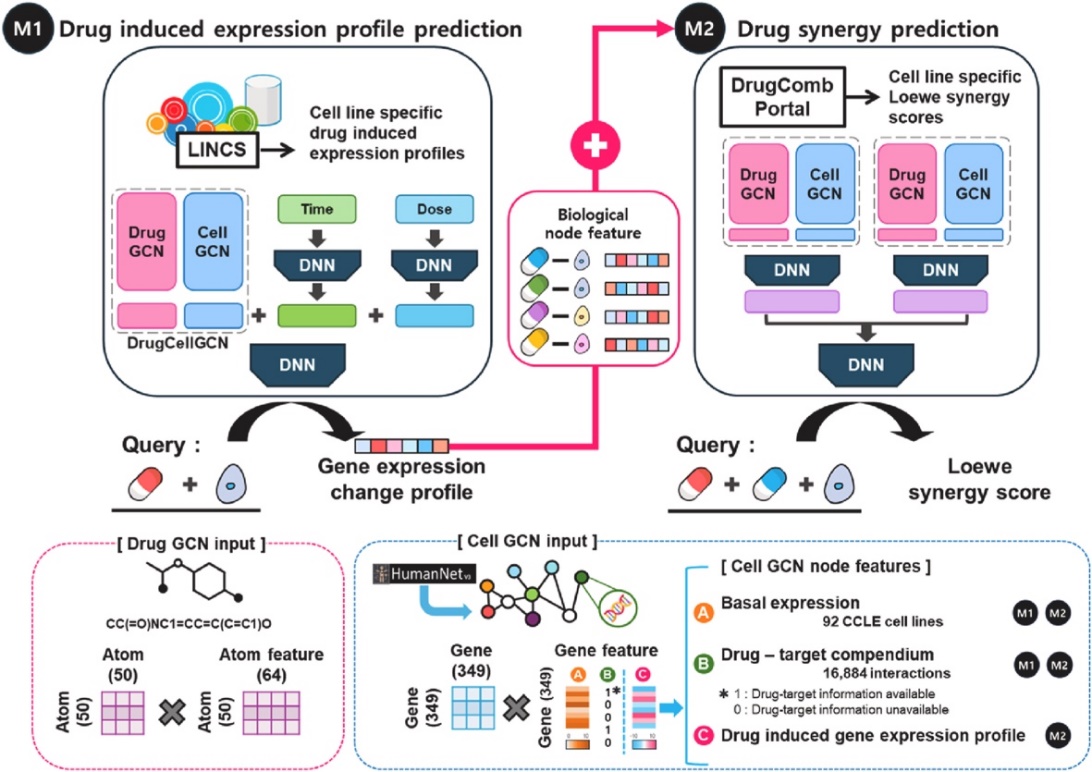
Figure 1. The overview of DRSPRING
Results
We first tested whether M1 and the combined model (M2) could predict cell-type-specific drug-drug combinations. The first module (M1) of DRSPRING learned LINCS gene expression profiles using 51,081 samples from 5,771 drugs and 86 cell lines. Biological networks, particularly HumanNet v3 with 349 genes, significantly improved prediction accuracy (MSE = 4.033, PCC = 0.572) compared to using other networks like STRING. Predicted versus experimental PCCs were high, with the top 10% averaging 0.9071, and accuracy varied by tissue type, driven by cell line homogeneity and data quantity, with the best performance seen in cell lines from pancreas, cervix, and breast. The second module (M2) of DRSPRING predicted Loewe scores for 294,444 drug combinations across 92 cell lines with a PCC of 0.710, showing variation across cancer subtypes. HER2-amplified breast cancers and Hodgkin lymphoma exhibited poor performance due to challenges in replicating tumor microenvironments
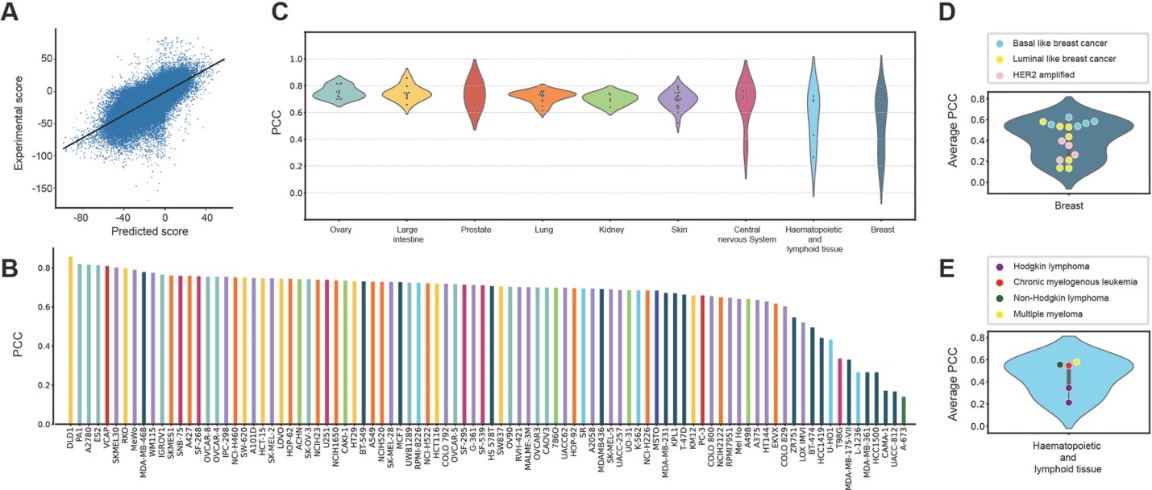
Figure 2. Performance of DRSPRING in predicting the Loewe synergy score. (A) The scatter plot of experimental and predicted Loewe scores. Linear regression yielded PCC = 0.7139. (B) The PCCs of experimental and predicted Loewe scores over drug pairs in each cell line. (C) Distribution of PCCs in each tissue type. (D) Violin plot of average PCCs in several breast cancer types. Each dot represents a cell line color-coded according to the cancer subtype (basal-like, luminal-like, and HER2-amplified). The average PCC was calculated over drug combinations in each cell line. (E) Violin plot of average PCCs in several haematopoietic and lymphoid tissues whose subtype was annotated in the CCLE.
To identify promising new drug pairs for specific cancer patients, we tested unknown drug combinations through DRSPRING. The model identified several high-scoring drug pairs, many of which were already in clinical trials or supported by literature. For example, combinations involving olaparib were found to be in clinical trials for multiple cancers, and several other candidates showed strong in vitro and in vivo evidence. However, many of the predictions should be validated in clinical, in vivo, or in vitro experiments, as they were based on data from different cancer or tissue types.
Significance DRSPRING is the first model to combine LINCS data with a GCN model for drug-drug-cell synergy prediction, addressing the challenge of data sparsity. We built an extensive drug-target database and provided predicted gene expression profiles that align closely with experimental data, enabling further applications like drug repurposing and side effect prediction. The method can be enhanced with recent deep learning techniques and has broad applicability, as it only requires gene expression data from public resources, making it a valuable tool for predicting drug synergy and developing combination therapies.
* Related Article
Jiyeon Han, Min Ji Kang, Sanghyuk Lee, DRSPRING: Graph convolutional network (GCN)-Based drug synergy prediction utilizing drug-induced gene expression profile,, Computers in Biology and Medicine, 174, 108436, 2024