본문
Enzymatic Synthesis of Self-assembled Dicer Substrate RNA Nanostructuresfor Programmable Gene Silencing
By Prof. Hyukjin Lee (hyukjin@ewha.ac.kr)
Department of Pharmacy
Self-assembled nucleic acid nanostructures are widely utilized for applications in drug delivery and molecular imaging. They offer unique advantages over the conventional nanoparticles made of polymers, inorganic, and carbon-based materials. These include the high-yield synthesis of molecularly identical nanostructures with precise control of shape and size.
Previously, large-scale production of DNA nanostructures was reported by rolling circle amplification (RCA) carrying multiple synthetic short interfering RNAs (siRNAs) into target cells. Although DNA nanostructures are particularly useful for delivering synthetic siRNAs into the cells, their major role is quite limited to provide physical and structural support for the region-specific hybridization of siRNAs within the systems.
Besides the DNA nanostructures, RNA nanostructures can provide bi-functional roles as a physical frame for intracellular delivery as well as a biological trigger to induce RNAi by itself. However, they required multiple long synthetic RNA strands to generate functional RNA nanostructures and it was very costly to prepare them in large quantities.
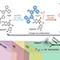
Herein, we report the large-scale preparation of RNA nanostructures through an enzymatic synthesis. As shown in Figure 1, three arm junction RNA nanostructures (Y-RNA) was synthesized in large quantities by the two-step reactions including isothermal rolling circle transcription (RCT) of DNA templates and site-specific cleavage of amplified RNA strands by RNase H treatment. Depending on the hybridization of helper strands, various Y-RNA with open or closed loop structure can be generated.
The resultant Y-RNA showed different gene silencing activity depending on existence of loop structure. Because Dicer only recognizes the 3′ end of ds-RNA, arms of Y-RNA with the closed loop structure cannot be processed by Dicer. The Y-RNA offered the simultaneous regulation of multiple target genes by controlling the ratio of delivered RNAi sequences in cells.
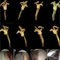
In addition, we have demonstrated structural requirements of Dicer substrate Y-RNA for enhanced RNAi effect. Compared to conventional siRNA, Y-RNA showed enhanced gene silencing at lower dose in vitro and in vivo. This result confirmed that endogenous Dicer processing of Y-RNA could facilitate the RISC formation. Collectively, our results showed the versatile potential of Dicer substrate RNA nanostructures and this approach may offer new strategies to develop future RNAi therapeutics
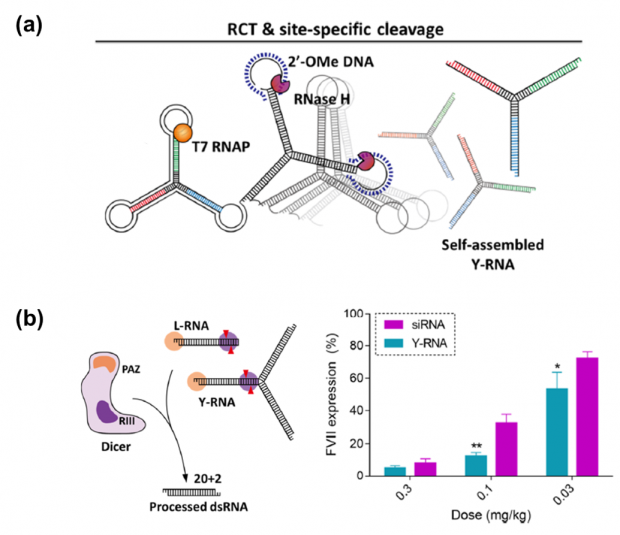
Figure 1. (a) Schematic illustration of enzymatic preparation of Y-RNA (b) Comparative evaluation of dose responsive FVII knockdown using siRNA andY-RNA (n = 3) (*, p-value ≤0.05; **, p-value ≤0.01).
* Related Article
[1] H. Lee, A.K. Lytton-Jean, Y. Chen, K.T. Love, A.I. Park, E.D. Karagiannis, A. Sehgal, W. Querbes, C.S. Zurenko, M. Jayaraman, C.G. Peng, K. Charisse, A. Borodovsky, M. Manoharan, J.S. Donahoe, J. Truelove, M. Nahrendorf, R. Langer, D.G. Anderson, Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery, Nature nanotechnology, 7 (2012), 389-393
[2] C.A. Hong, B. Jang, E.H. Jeong, H. Jeong, H. Lee, Self-assembled DNA nanostructures prepared by rolling circle amplification for the delivery of siRNA conjugates, Chemical communications, 50 (2014) 13049-13051.
[3] B. Jang, B. Kim, H. Kim, H. Kwon, M. Kim, Y. Seo, M. Colas, H. Jeong, E.H. Jeong, K. Lee, H. Lee, Enzymatic Synthesis of Self-assembled Dicer Substrate RNA Nanostructures for Programmable Gene Silencing, Nano letters, 18 (2018) 4279-4284.
* Video Clip
Development of RNA nanostructures for enhanced RNAi potency