본문
Rpd3L HDAC links H3K4me3 to transcriptional repression memory
By Prof. TaeSoo Kim (tskim@ewha.ac.kr)
Department of Life Science
In their natural growth conditions, cells are exposed to rapidly changing environments and must accordingly reprogram their gene expression patterns for cellular differentiation, development, and adaptation. In addition, they are often re-exposed to the same or different stimuli and can rapidly re-induce the genes required for cellular functions. This response is known as ‘transcriptional memory’ and increases the kinetics of reactivation. Although cells must repress unnecessary genes upon environmental changes, whether a gene remembers its previous transcriptionally inactive state remains unclear. In this study, we show that transcriptional repression of ∼540 genes in yeast occurs much more rapidly if the genes have been previously repressed during carbon source shifts. This novel transcriptional response has been termed transcriptional repression memory (TREM).
TREM may be induced by rapid dissociation of activators from promoters. Alternatively, repressors that bind to active promoters could induce TREM. A conserved histone deacetylase, Rpd3 functions as a transcriptional repressor and is the catalytic subunit of at least two distinct HDACs. Rpd3 large (Rpd3L) HDAC includes more than 10 subunits and presumably deacetylates histones at promoter regions. Genome-wide chromatin immunoprecipitation-DNA microarray (ChIP-ChIP) analyses revealed that Rpd3L associates with actively transcribed genes. Consistent with this data, the loss of Rpd3L increases histone acetylation at active promoters. In contrast, Rpd3 small (Rpd3S) HDAC primarily deacetylates histones within coding regions to suppress initiation from cryptic promoters, and histone exchange within coding regions, and RNA Pol II elongation.
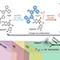
Interestingly, Rpd3L targeted to active promoters induces TREM. Mutants for Rpd3L but not for Rpd3S exhibit increased acetylation at active promoters and delay TREM significantly. Surprisingly, the interaction between H3K4me3 enriched at active promoters and Rpd3L via the Pho23 PHD finger is critical to promote histone deacetylation and TREM by Rpd3L. Therefore, we propose that an active mark, H3K4me3 enriched at active promoters, instructs Rpd3L HDAC to induce histone deacetylation and TREM.
In this study, we show, for the first time, that many yeast genes exhibit ‘memory’ of their preceding transcriptional inactive state, and this phenomenon is termed TREM. TREM seems to be biologically important because enhancement of transcriptional repression via TREM may save cellular energy and resources including transcription machinery by rapidly turning off unnecessary genes for cellular functions. Recent studies have suggested that transcriptional/epigenetic memory plays an important role in innate immunity. Innate Lymphoid Cells (ILCs) mediate non-specific responses to allergens and were found to be hyperactivated upon re-exposure to the same or unrelated allergens. Importantly, a small portion of previously trained cells will become naïve cells once the allergen is eliminated. Repression of unnecessary or hyperactivated genes by TREM is likely important to generate naïve cells from trained cells.
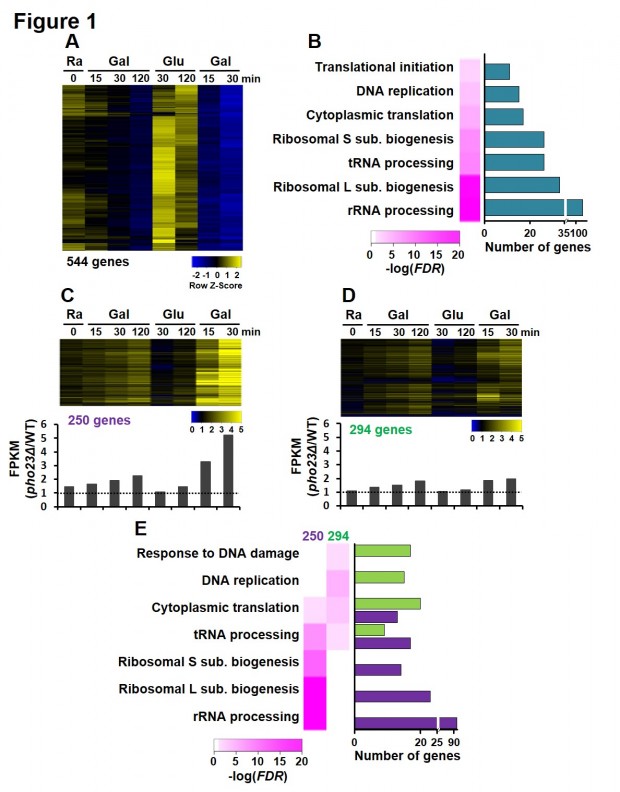
Figure 1. Rpd3L is required for TREM. (A) Expression profiles of 544 TREM genes. RNA samples from PHO23 and pho23Δ strains were analyzed by strand-specific RNA sequencing. The TREM genes identified in PHO23 strains are visualized. (B) GO analysis of TREM genes. Many TREM genes are involved in ribosomal RNA processing and ribosomal large or small subunit biogenesis. (C) TREM of 250 genes is significantly delayed in pho23Δ cells. The ratio of transcript levels for 250 genes in pho23Δ over those in PHO23 are visualized. Bottom panel shows the averaged expression ratios at each time point. A strong difference in transcript levels is seen during the second galactose pulse. (D) TREM of 294 genes is not regulated by Pho23. The ratio of transcript levels is shown as in (C). (E) GO analysis of Pho23-sensitive and Pho23-insensitive genes. Whereas Pho23-sensitive genes are enriched for rRNA processing and ribosomal subunit biogenesis, Pho23-insensitive genes are mostly involved in DNA replication and DNA damage response.
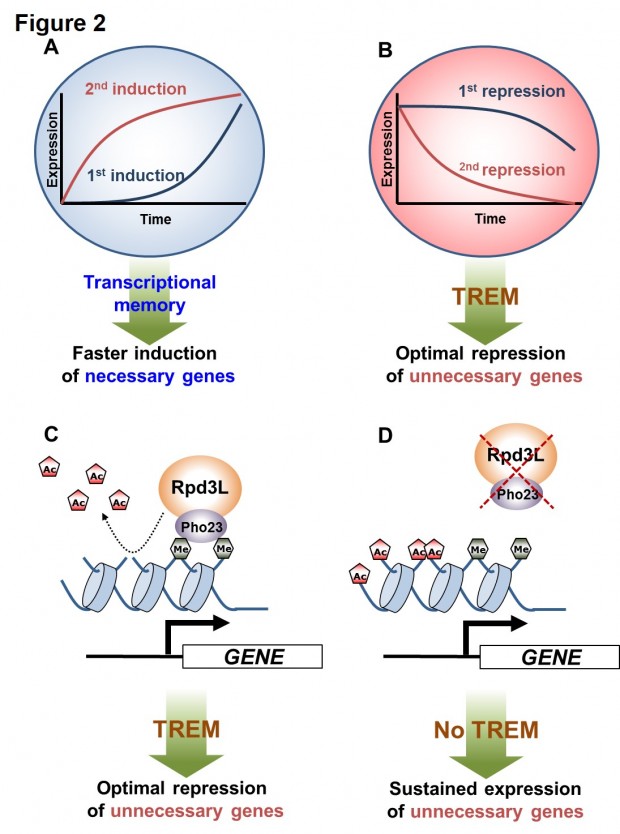
Figure 2. Transcriptional memory and models for regulation of TREM by Rpd3L. (A) When cells are exposed to a stimulus, the genes necessary for cellular functions are induced (first induction). Transcriptional memory upon re-exposure to the same stimulus significantly increases the rate of gene activation (second induction). (B) In contrast, unnecessary genes are repressed by a stimulus (first repression). TREM upon re-exposure to the same stimulus promotes optimal gene repression (second repression). (C) Hypoacetylation mediated by the Rpd3L-H3K4me3 interaction results in optimal repression of unnecessary genes by TREM. (D) Hyperacetylation by the loss of Rpd3L or by the loss of the Pho23 PHD finger-H3K4me3 interaction result in sustained expression of unnecessary genes.
* Related Article
Bo Bae Lee et al., Rpd3L HDAC links H3K4me3 to transcriptional repression memory, September 2018, Nucleic Acids Research, Vol 46, 8261-8274